Overview
The problem
WHO has ranked antimicrobial resistance (AMR) among the top ten global threats to public health. In 2019, Italy experienced over 10,000 deaths, this set a sad record of Italy as country with the highest number of AMR-associated deaths in the European Community. Due to their innate AMR and severe pathogenicity, of particular interest are the so called ESKAPE bacteria, namely: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species.
The speed of onset and spreading of new AMR strains outclasses the development of new antibiotic molecules by the pharmaceutical industry. Therefore, it is necessary to devise new therapeutic approaches to counter AMR infections.
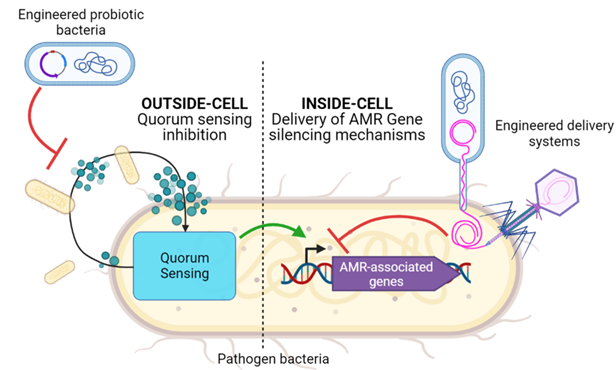
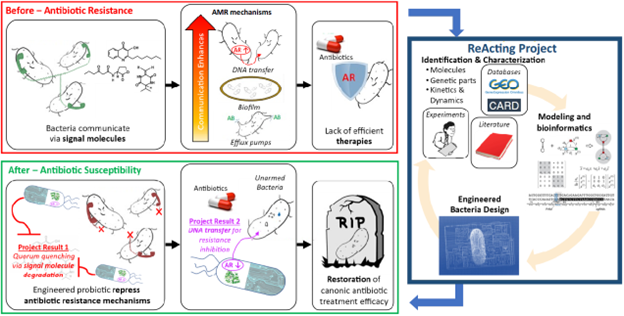
Multiple synergetic mechanisms are involved in AMR, operating both at the single bacterium level, via expression of AMR-promoting genes, and at bacterial community level, where bacterium-to-bacterium communications enable coordinated population actions. The latter includes quorum sensing (QS) signaling, where microbes coordinate AMR-promoting actions, such as biofilm formation and exchange of DNA, by means of diffusible signal molecules.
The idea
By applying Systems and Synthetic Biology (SB) techniques, this project intends to create engineered bacteria tackling AMR both at single bacterium and at population level. At single bacteria level, we will engineer therapeutic bacteria that horizontally transfer a CRISPRi-based repressor for AMR-associated gene expression. At population level, the engineered bacteria will interfere with QS mechanisms, thus restoring sensitivity to antibiotic molecules.
As a case study, we will apply this approach to fight some of the foremost clinically frightening ESKAPE pathogens: Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa.
Our research group includes bioengineers, control theorists, clinical microbiologists and bioinformaticians, covering aforementioned skills, facilities and technologies, to converge in the synergy necessary to successfully complete the project and to develop novel therapies. This highly technological and interdisciplinary context lead to the prospect of creating a center of knowledge and excellence, with international attractiveness and possibly economical interest in Veneto region.
Objectives
The approach
The aim of this project is to propose a novel therapeutic paradigm based on systems and synthetic biology approaches. By gathering information from literature, genome expression databases and model driven experimental characterizations, we aim to implement a mathematical model that recapitulates the main AMR mechanism of those bacteria and the relationship with QS mechanisms, allowing the simulation of possible therapeutic scenarios.
Driven by this model, we plan to rationally design and engineer a probiotic bacterial strain that can interact with the surrounding bacteria populations, inducing a decrease in AMR in terms of minimal inhibitory concentration, thus enabling the eradication of the pathogens through canonical antibiotic administration.
The solution
Specifically, the primary goals can be summarized as:
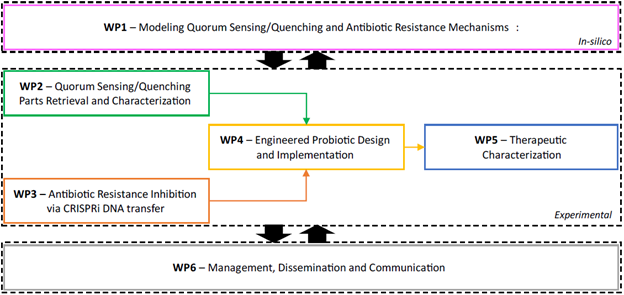
- Develop a model recapitulating AMR, QS, and their relationship [WP1].
- Retrieve, design, and implement genetic parts and circuits to inhibit AMR at two levels:
- Outside-cell: inhibiting QS via signal molecules degradation/sequestration [WP2]
- Inside-cell: transferring AMR-associated genes repressors via conjugation of CRISPRi systems [WP3].
- Engineer, optimize and validate a therapeutic probiotic strain to perform the required tasks [WP4].
- Obtain pre-clinical validation in vitro (pathogen-probiotic co-culture) and in vivo (murine model) [WP5].
- Increase public opinion and stakeholders’ interest in this novel high-potential field, possibly starting a market analysis and patenting the developed stain [WP6].
The whole project will be sized and focused on the most representative bacterial species found in Padua University Hospital’s cohort of patients. As such, the project will provide a proof of concept to be used as a showroom to involve pharma/biotech companies on the possible advantages of exploiting engineered organisms as an adjuvant or even alternative solution for those cases and pathologies where the lack of efficient therapies hampers the treatments. Lastly, it is worth to notice that the modularity of the project allows the production of several “self-standing” impacting results (e.g., genetic parts and circuits, biological parameters, and mathematical relationships) to be shared with the scientific community.
Publications
- ECCB 2023 – Bellato M, et al. “Bactlife – A Dash GUI to simulate bacterial communities’ evolution via agent-based modeling” –Conference Paper;
- Bernabè G, et al. “A novel phenolic derivative inhibits AHL-dependent quorum sensing signaling in Pseudomonas aeruginosa”, Frontiers in Pharmacology, 13, 2022, link;
- GNB 2023 – Gaetan E, et al. “Modeling metabolic overload effects in bacterial growth rate in synthetic biology” – Conference paper accepted for oral presentation;
- GNB 2023 – Bellato M, et al. “Simulating microbial communities’ evolution via Agent base modelling: a Python tool” – Conference paper;
- GNB 2023 – Bernardi A, et al. “Automated control of bioreactors: An hardware-in-the-loop proof of concept test towards an experimental facility” – Conference paper;
- GNB 2023 – Cimolato C, et al. “Uncovering quorum sensing and quenching structural properties: a systems biology approach” – Conference paper;
- BITS 2022 – Bellato M, et al. “Implementation of a Python simulator for microbial communities’ evolution via agent-based modeling” – Conference paper;
- BBCC 2022 – Bellato M, et al. “An agent-based simulator for microbial communities’ evolution” – Conference paper accepted for oral presentation, winner of the best oral prize.